Topic 10.2: Inheritance
In the Inheritance unit we will learn that heredity, or the continuity of traits between parent and offspring, is powered by the physical transmission of DNA between cells during reproduction. In this unit, you will learn about the origins of our modern understanding of heredity, as well as the basic rules that determine how parents’ traits determine the traits of their offspring.
This unit will last 3 school days
This unit will last 3 school days
Essential idea:
- Genes may be linked or unlinked and are inherited accordingly.
Nature of science:
- Looking for patterns, trends and discrepancies—Mendel used observations of the natural world to find and explain patterns and trends. Since then, scientists have looked for discrepancies and asked questions based on further observations to show exceptions to the rules. For example, Morgan discovered non-Mendelian ratios in his experiments with Drosophila. (3.1)
- Describe the trends and discrepancies that led Morgan to propose the idea of linked genes.
- Describe the trends and discrepancies that led Morgan to propose the idea of linked genes.
Understandings
10.2 U 1 Gene loci are said to be linked if on the same chromosome. (Oxford Biology Course Companion page 448).
- Define autosome and sex chromosome.
- Describe what makes genes “linked.”
Linkage refers to genes that are located on the same chromosome. Linked genes tend to be inherited together and the extent of crossing over depends on how close together they are on the chromosome. Linkage generally reduces the variety of offspring that can be produced. In genetic crosses, linkage is indicated when a greater proportion of the offspring resulting from a cross are of the parental type (than would be expected if the alleles were assorting independently). If the genes in question had been on separate chromosomes, there would have been more genetic variation in the gametes and therefore in the offspring.
10.2 U 2 Unlinked genes segregate independently as a result of meiosis. (Oxford Biology Course Companion page 445).
- State the difference between independent assortment of genes and segregation of alleles.
- Describe segregation of alleles and independent assortment of unlinked genes in meiosis.
Unlinked genes are on different chromosomes. At Metaphase 1, each pair of homologous chromosomes line up at the equator of the spherical cell. For each homologous pair, the chromosomes assort to one side or the other. The alignment of any pair of homologous chromosomes is independent of all the other pairs. Thus, the mixture of unlinked genes in gametes is random.
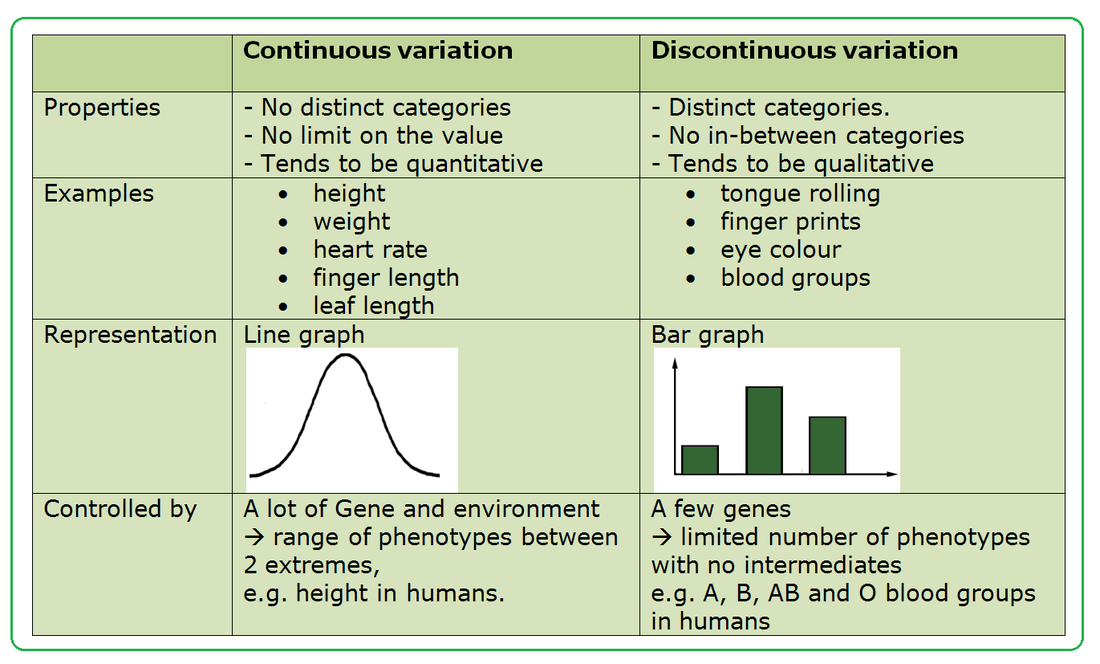
10.2 U 3 Variation can be discrete or continuous.
Discontinuous variation This is where individuals fall into a number of distinct classes or categories, and is based on features that cannot be measured across a complete range. You either have the characteristic or you don't. Blood groups are a good example: you are either one blood group or another - you can't be in between, same goes for gender. Such data is called discrete (or categorical) data. Chi-squared statistical calculations work well in this case.
In continuous variation there is a complete range of measurements from one extreme to the other. Height is an example of continuous variation - individuals can have a complete range of heights, for example, 1.6, 1.61, 1.62, 1.625 etc metres high.
In continuous variation there is a complete range of measurements from one extreme to the other. Height is an example of continuous variation - individuals can have a complete range of heights, for example, 1.6, 1.61, 1.62, 1.625 etc metres high.
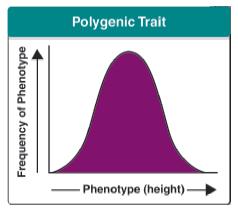
10.2 U 4 The phenotypes of polygenic characteristics tend to show continuous variation (Oxford Biology Course Companion page 450).
- Explain polygenetic inheritance using an example of a two gene cross with codominant alleles.
- State that a normal distribution of variation is often the result of polygenic inheritance.
- State example human characteristics that are associated with polygenic inheritance.
Continuous variation is the combined effect of many genes (known as polygenic inheritance) and is often significantly affected by environmental influences. Milk yield in cows, for example, is determined not only by their genetic make-up but is also significantly affected by environmental factors such as pasture quality and diet, weather, and the comfort of their surroundings.
Polygenic traits are controlled by two or more than two genes (usually by many different genes) at different loci on different chromosomes. These genes are described as polygenes. Examples of human polygenic inheritance are height, skin colour and weight. Polygenes allow a wide range of physical traits. For instance, height is regulated by several genes so that there will be a wide range of heights in a population.
Polygenic traits are controlled by two or more than two genes (usually by many different genes) at different loci on different chromosomes. These genes are described as polygenes. Examples of human polygenic inheritance are height, skin colour and weight. Polygenes allow a wide range of physical traits. For instance, height is regulated by several genes so that there will be a wide range of heights in a population.
10.2 U 5 Chi-squared tests are used to determine whether the difference between an observed and expected frequency distribution is statistically significant (Oxford Biology Course Companion page 453)
- State the two possible hypotheses of a statistical test.
- Calculate the chi square value to determine the significance of differences between the observed and expected results of a genetic cross.
- Determine the degrees of freedom and critical value for the chi-square test.
- Draw a conclusion of significance by comparing the calculated and critical chi-square values.
A chi-square test is a statistical test that can be used to determine whether observed frequencies are significantly different from expected frequencies. These statistical tests enable us to compare observed and expected frequencies empirically and to decide if the results we see are statistically significant. Statistical significance in this case implies that the differences are not due to chance alone, but instead may be caused by other factors at work.
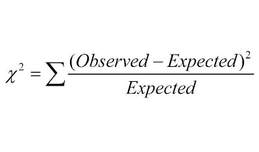
What it basically means is the sum of the (observed minus the expected) squared, divided by the expected. This value is then compared to a critical value from a chi-squared table (similar to a t-table) to determine if the numbers we see are due to random factors. This is useful in determining if the results from a dihybrid genetic cross are due to independent assortment.
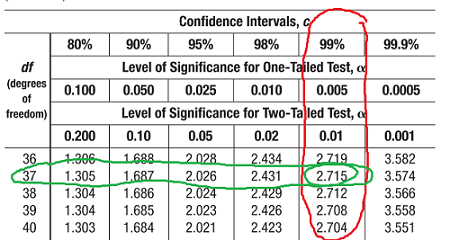
- Degree of freedom - the number of independent values or quantities that can be assigned to a statistical distribution.
- To find the degree of freedom = n - 1, where n is the number of data points
- A critical value is the number you compare your own result to (critical values would be provided in a test).
- A confidence interval does not quantify variability. A 95% confidence interval is a range of values that you can be 95% certain contains the true mean of the population.
(Ignore the circled row and column)
If your OWN value for something is greater than the critical value, you REJECT the null hypothesis and ACCEPT the experimental hypothesis.
If you OWN value for something is less than the critical value, you ACCEPT the null hypothesis and REJECT the experimental hypothesis.
If your OWN value for something is greater than the critical value, you REJECT the null hypothesis and ACCEPT the experimental hypothesis.
If you OWN value for something is less than the critical value, you ACCEPT the null hypothesis and REJECT the experimental hypothesis.
Application
10. 2 A 1 Morgan’s discovery of non-Mendelian ratios in Drosophila. (Oxford Biology Course Companion page 446).
- Determine possible allele combinations in gametes for crosses involving two genes.
- Use correct notation to depict a dihybrid cross between two unlinked genes.
- Construct a Punnett square to show the possible genotype and phenotype outcomes in a dihybrid cross.
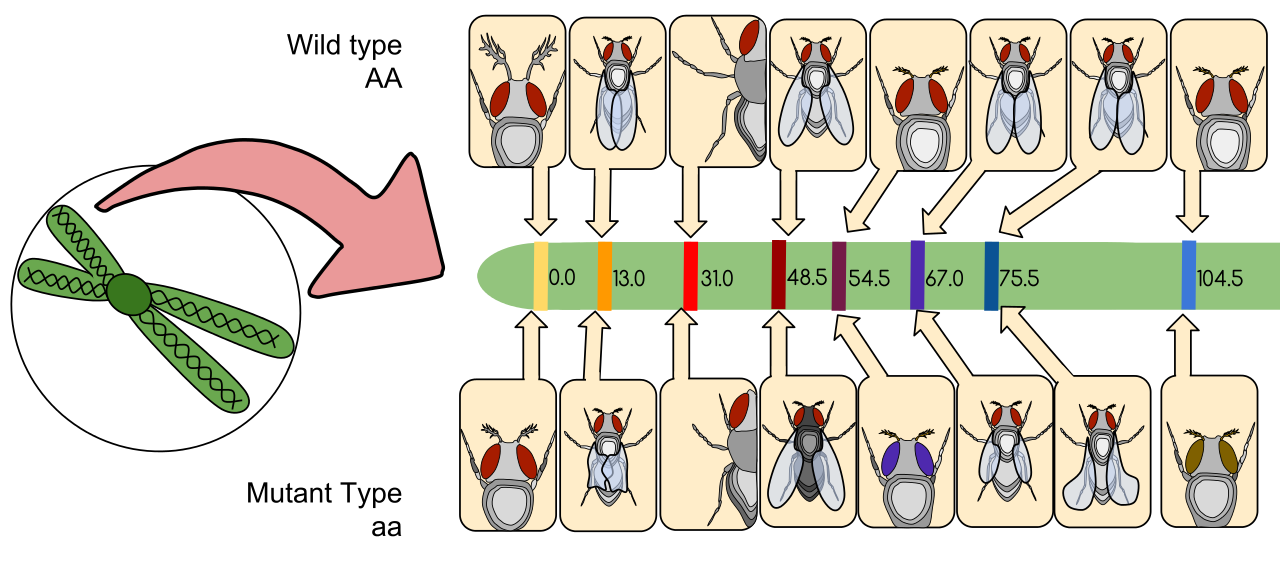
- Male fly with white eyes (flies usually have red eyes) was crossed with a female fly with red eyes (a wild type)
- The offspring of the first generation were red eyed
- Males and females of the first generation were crossed with each other
- The white-eye trait reappeared in the expected 3:1 Mendelian ratio for a recessive trait. However only the males had white eyes
- This suggested that the white-eye trait is carried on the X chromosome
- Crossing white-eyed males and red-eyed females from the second generation produced equal numbers of offspring with each eye colour
- Males have white eyes when they inherit the mutant gene on the X chromosome from their mother. Females only show the trait if they inherit mutant genes on both X chromosomes. This became known as "sex linkage
10.2 A 2 Completion and analysis of Punnett squares for dihybrid traits. (Oxford Biology Course Companion page 448).
- Describe how Morgan discovered relationship between eye color and sex in Drosophila.
Linked genes that have undergone recombination can be distinguished from unlinked genes via a test cross because the frequency of the recombinant genotypes will always be less than would occur for unlinked genes (crossing over does not happen every time)
For example:
For example:
- Heterozygous test cross of unlinked genes = 1 : 1 : 1 : 1 phenotypic ratio
- Heterozygous test cross of linked genes = 1 : 1 : 0.1 : 01 phenotypic ratio (uncommon phenotypes are recombinants)
- A dihybrid cross is a cross between two individuals that shows the inheritance of two different genes at the same time; usually involving unlinked autosomal genes.
- Note: The following example contains two unlinked genes, which means the genes are on different chromosomes. This means they follow Mendel’s law of independent assortment.
Dihybrid Cross Example
- Since almost all examples look at Mendel’s pea plants, for this example we will look at two traits in cats; hair length and color.
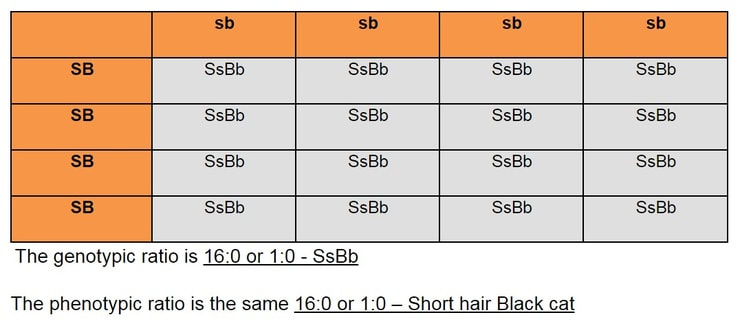
- If the question stated that short hair (S) is dominant over long hair (s) and black fur (B) is dominant over white fur (b), what would be the genotypic and phenotypic ratio of a cross between a short hair black cat that is homologous for these traits and a long hair white cat?
Next, write out the possible gametes: SB, SB, sb and sb or simply just SB and sb
10.2 A 3 Polygenic traits such as human height may also be influenced by environmental factors.(Oxford Biology Course Companion page 450).
- Outline two example environmental factors that can influence phenotypes.
- Compare continuous to discrete variation.
When one gene controls the expression of a trait, the number of phenotypes that are expressed is limited to the dominant phenotype or recessive phenotype. If there is co-dominance, this adds another possible phenotype that can be expressed.
With polygenic inheritance when two or more genes control the expression of a phenotype many possible phenotypes can exist. As the amount of genes that control one trait increase, the number of phenotypes increases to a point where it is impossible to determine the genotype by just observing the phenotype. Each additional gene has an additive affect, increasing the phenotypes. This is called continuous variation.
For example, people’s skin color varies dramatically around the world, between people of different races and within the same race. The multiple genes affect the intensity of the pigments in the skin. Another example is human height, which varies from person to person within the same race, and varies between different races. Height shows continuous variation.
If you graphed the frequency of the occurrence of different phenotypic variations in a population, there should be a normal distribution. Continuing with human height, there will be some really tall people and some really short people, but the majority of people will be average height (normal distribution). As the number of genes that control a certain trait increases, the closer the distribution of the phenotypes represents a normal distribution.
With polygenic inheritance when two or more genes control the expression of a phenotype many possible phenotypes can exist. As the amount of genes that control one trait increase, the number of phenotypes increases to a point where it is impossible to determine the genotype by just observing the phenotype. Each additional gene has an additive affect, increasing the phenotypes. This is called continuous variation.
For example, people’s skin color varies dramatically around the world, between people of different races and within the same race. The multiple genes affect the intensity of the pigments in the skin. Another example is human height, which varies from person to person within the same race, and varies between different races. Height shows continuous variation.
If you graphed the frequency of the occurrence of different phenotypic variations in a population, there should be a normal distribution. Continuing with human height, there will be some really tall people and some really short people, but the majority of people will be average height (normal distribution). As the number of genes that control a certain trait increases, the closer the distribution of the phenotypes represents a normal distribution.
Skills
10.2 S 1 Calculation of the predicted genotypic and phenotypic ratio of offspring\ of dihybrid crosses involving unlinked autosomal genes. (Oxford Biology Course Companion page 447).
- Determine the predicted genotype and phenotype ratios of F1 and F2 offspring of dihybrid crosses.
See 10.2.A2 Above
10.2 S 2 Identification of recombinants in crosses involving two linked genes. (Oxford Biology Course Companion page 451).
- Use correct notation to show alleles of linked genes.
- Construct a Punnett square to show the possible genotype and phenotype outcomes in a dihybrid cross involving linked genes.
- Explain how crossing over between linked genes can lead to genetic recombinants.
In a cross between two true breeding or homozygous parents the following would be the genotypes of the cross: TTBB x ttbb.
On an exam if the genes are linked the genotypes would be represented as follows:
Explanation of Recombinants
The genotypes represented on the right show that T and B are on the same chromosome with one pair on the maternal chromosome and one pair on the paternal chromosome
TB, Tb, and tB would now be the new linked alleles for the next generation
On an exam if the genes are linked the genotypes would be represented as follows:
Explanation of Recombinants
The genotypes represented on the right show that T and B are on the same chromosome with one pair on the maternal chromosome and one pair on the paternal chromosome
- The horizontal line represents the homologous chromosomes
- The same can be said about the alleles t and b
- The resulting cross should show that all the offspring are heterozygous for the traits
- As the genes are linked T and B will go into the same gamete as will t and b, resulting in the heterozygous TtB
- If we now crossed the two heterozygous offspring TtBb the cross would be represented by the symbols on the right
- Since the genes are linked T and B and t and b chromatids should be inherited together
- The resulting gametes would contain TB and tb alleles, therefore the majority of the offspring will be TTBB, TtBb and ttbb
- This does not follow Mendel’s law off independent assortment because the genes are on the same chromosomes
- However, crossing over can occur between the non-sister chromatids during prophase I of meiosis resulting in offspring with different genotypes
- The gametes Tb and tB could also occur with crossing over, resulting in the recombinant offspring TtBB and TTBb
- Recombinants have different alleles combinations than either of the parents
TB, Tb, and tB would now be the new linked alleles for the next generation
10.2 S 3 Use of a chi-squared test on data from dihybrid crosses. (Oxford Biology Course Companion page 453).
- Calculate a chi-square value to compare observed and expected results of a dihybrid genetic cross.
- Using the df and critical chi-square value, determine if there is a significant difference between observed and expected results of a dihybrid cross.
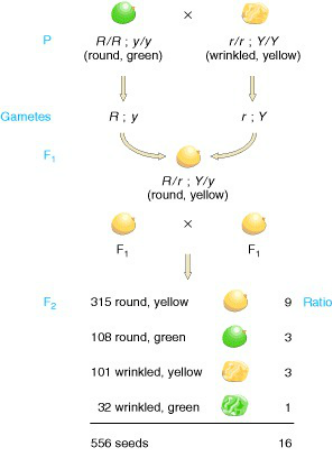
For an example, use Mendel’s results from his pea plant crosses.
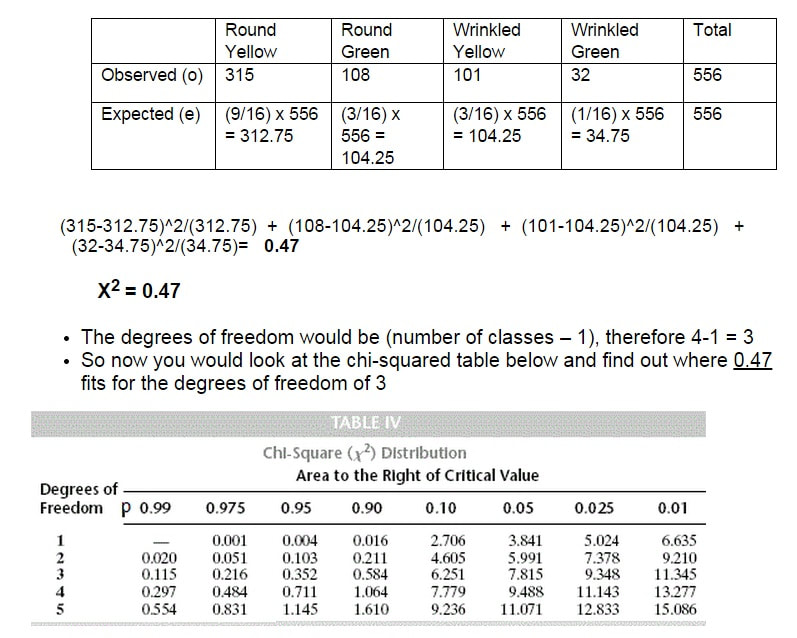
When he did a dihybrid cross between two heterozygotes RrYy x RrYy, the expected phenotypic ratio due to independent assortment would be 9:3:3:1. Look at the chart below to see his actual results.
When he did a dihybrid cross between two heterozygotes RrYy x RrYy, the expected phenotypic ratio due to independent assortment would be 9:3:3:1. Look at the chart below to see his actual results.
The Ho (null hypothesis) would be the results are due to independent assortment and the Ha (alternative hypothesis) would be that the alleles do not assort independently and the results are due to gene linkage.
As you can see from the table above the critical value at the 0.05 level of significance is 7.815.
What the p-value of 0.05 or 5% indicates is the probability of getting the results you did (or more extreme results) given that the null hypothesis is true.
If the calculated value is above or equal to 7.815 then we reject the Ho and accept the HA that the alleles in question are linked. However, since our chi-squared value of 0.47 is significantly less than this value (it has a p-value of 0.90 or 90%) we accept the Ho that the results that Mendel saw were due to independently assortment of the alleles.
NOTE: Alleles are usually shown side by side in dihybrid crosses, for example, TtBb. In representing crosses involving linkage, it is more common to show them as vertical pairs, for example:
What the p-value of 0.05 or 5% indicates is the probability of getting the results you did (or more extreme results) given that the null hypothesis is true.
If the calculated value is above or equal to 7.815 then we reject the Ho and accept the HA that the alleles in question are linked. However, since our chi-squared value of 0.47 is significantly less than this value (it has a p-value of 0.90 or 90%) we accept the Ho that the results that Mendel saw were due to independently assortment of the alleles.
NOTE: Alleles are usually shown side by side in dihybrid crosses, for example, TtBb. In representing crosses involving linkage, it is more common to show them as vertical pairs, for example:
Key Terms
|
genotypes
F1 generation linkage group discontinuous degrees of freedom meiosis codominant polygenic crosses Drosophila gene loci |
autosome
gametes F2 generation Mendelian ratios sex-linked variation Chi-square genotype |
haploid
Mendel linkage genotype polygenes un-linked genes sex chromosome Pascal’s triangle Punnett Square df |
dihybrid cross
chromosome monohybrid independent assortment discrete variation continuous variation polygenic inheritance distribution frequency genetic recombinants |
parental generation
linked discrete chi-square segregation phenotype allele Morgan segregation |
PowerPoint and Notes on Topic 10.1 by Chris Payne
Your browser does not support viewing this document. Click here to download the document.
Your browser does not support viewing this document. Click here to download the document.
Correct use of terminology is a key skill in Biology. It is essential to use key terms correctly when communicating your understanding, particularly in assessments. Use the quizlet flashcards or other tools such as learn, scatter, space race, speller and test to help you master the vocabulary.
Useful Links
In The News
Study of short Peruvians reveals new gene with a major impact on height - Science, May 2018
New gene variants reveal the evolution of human skin color - Science, Oct 2017
TOK
- The law of independent assortment was soon found to have exceptions when looking at linked genes. What is the difference between a law and a theory in science?
Video Clips
Each father and mother pass down traits to their children, who inherit combinations of their dominant or recessive alleles. But how do we know so much about genetics today?
Hank and his brother John discuss heredity via the gross example of relative ear wax moistness.
For all of human history, we've been aware of heredity. Children look like their parents. But why? When Gregor Mendel pioneered the field of genetics, he began to try to answer that question, and now that we know about chromosomes, all of his work will make a lot of sense. Let's take a look at Mendel's work, his laws of segregation and independent assortment, and try to understand what these mean for genetics and inheritance
Paul Andersen introduces the Punnett Square as a a powerful tool in genetic analysis. He tries to address major misconceptions that students have when use a Punnett Square. He gives a number of examples of monohybrid crosses and one example of a dihybrid cross. The square is a simple tool that uses the outcome of meiosis to determine possible offspring in a cross.
This video will show how to set up and solve everyone's favorite 16 square Punnett square
Explore inheritance when carried on the X chromosome with the Amoeba Sisters!
Discover more types of non-Mendelian inheritance such as incomplete dominance and codominance with the Amoeba Sisters!
In this video Paul Andersen explains the elements in the Hardy-Weinberg equation; including the allele frequency and phenotype frequency. He begins with a brief explanation of phenotypes and genotypes. A sample problem is worked out and another problem is included.
Paul Andersen shows you how to solve simple Hardy-Weinberg problems. He starts with a brief description of a gene pool and shows you how the formula is derived. He then shows you how to solve a couple of sample problems.
Paul Andersen shows you how to calculate the ch-squared value to test your null hypothesis. He explains the importance of the critical value and defines the degrees of freedom. He also leaves you with a problem related to the animal behavior lab.