Topic 7.1 DNA Structure and Replication
In the DNA Structure and Replication we will extend our knowledge on the DNA Structure from Topic 2.6. This unit will also focus on the individuals responsible for the discovery of the DNA helix and proving that DNA is the genetic code.
This unit will last 3 school days.
This unit will last 3 school days.
Essential idea:
- The structure of DNA is ideally suited to its function.
Nature of science:
- Making careful observations—Rosalind Franklin’s X-ray diffraction provided crucial evidence that DNA is a double helix. (1.8)
- Describe Rosalind Franklin’s role in the elucidation of the structure of DNA.
- Describe Rosalind Franklin’s role in the elucidation of the structure of DNA.
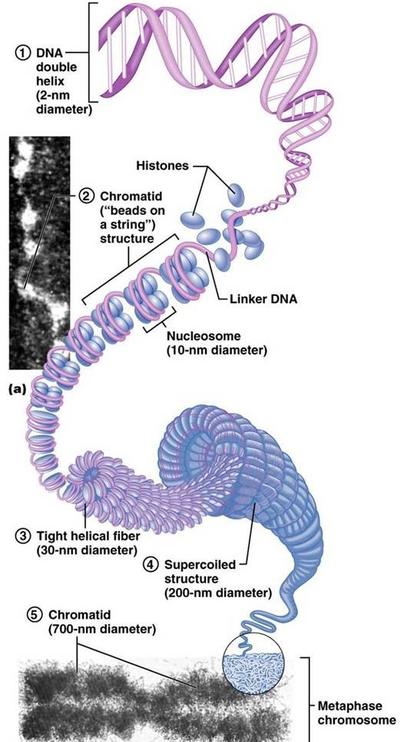
7.1 U1 Nucleosomes help to supercoil the DNA.
- Draw and label the structure of a nucleosome, including the H1 protein, the octamer core proteins, linker DNA and two wraps of DNA.
- Explain the levels of supercoiling (DNA→ nucleosome → beads on a string → 30nm fiber → unreplicated interphase chromosome → replicated metaphase chromosome).
|
DNA is arranged in chromosomes in eukaryotes. Each chromosome contains one very long DNA molecule.
The nucleosomes arrange into coils, and in preparation for nuclear division, the coiled structure coils up again, to form a supercoiled chromosome.. Nucleosomes are important for the safe storage of DNA. They also play an important role in the regulation of transcription. |
7.1 U 2 DNA structure suggested a mechanism for DNA replication. (Details of DNA replication differ between prokaryotes and eukaryotes. Only the prokaryotic system is expected) (Oxford Biology Course Companion page 347).
- Outline the features of DNA structure that suggested a mechanism for DNA replication.
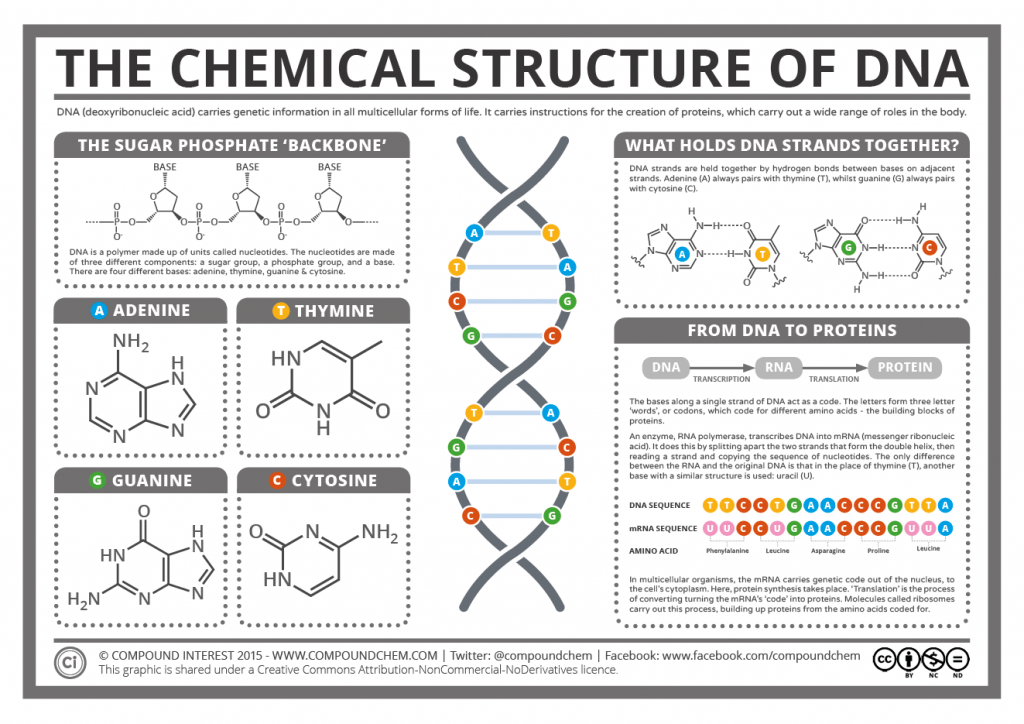
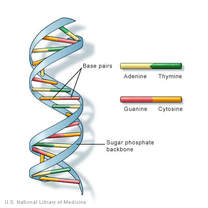
DNA is double stranded and shaped like a ladder, with the sides of the ladder made out of repeating phosphate and deoxyribose sugar molecules covalently bonded together. Each deoxyribose molecule has a phosphate covalently attached to a 3’ carbon and a 5’ carbon. The phosphate attached to the 5’ of one deoxyribose molecule is covalently attached to the 3’ of the next deoxyribose molecule forming a long single strand of DNA known as the DNA backbone. DNA strands run antiparallel to each other with one strand running in a 5’ to 3’ direction and the other strand running 3’ to 5’ when looking at the strands in the same direction.
- The rungs of the ladder contain two nitrogenous bases (one from each strand) that are bonded together by hydrogen bonds.
- Since these two strands are anti-parallel replication occurs in different directions on the DNA strand
- Purines are two ring nitrogenous bases and pyrimidines are single ring nitrogenous bases.
- The nitrogenous bases match up according the Chargaff’s Rules in which adenine (purine) always bonds to thymine (pyrimidine), and guanine (purine) always bonds with cytosine (pyrimidine).
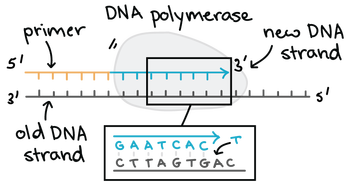
7.1 U 3 DNA polymerases can only add nucleotides to the 3’ end of a primer. (Oxford Biology Course Companion page 349).
- Compare replication on the the leading strand and the lagging strand of DNA.
- Explain why replication is different on the leading and lagging strands of DNA.
- Outline the formation of Okazaki fragments on the lagging strand.
The DNA polymerases are enzymes that create DNA molecules by assembling nucleotides, the building blocks of DNA. These enzymes are essential to DNA replication and usually work in pairs to create two identical DNA strands from one original DNA molecule. During this process, DNA polymerase “reads” the existing DNA strands to create two new strands that match the existing ones.
Every time a cell divides, DNA polymerase is required to help duplicate the cell’s DNA, so that a copy of the original DNA molecule can be passed to each of the daughter cells. In this way, genetic information is transmitted from generation to generation.
Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its tightly woven form. This opens up or “unzips” the double stranded DNA to give two single strands of DNA that can be used as templates for replication.
DNA polymerase adds new free nucleotides to the 3’ end of the newly-forming strand, elongating it in a 5’ to 3’ direction. However, DNA polymerase cannot begin the formation of this new chain on its own and can only add nucleotides to a pre-existing 3'-OH group. A primer is therefore needed, at which nucleotides can be added. Primers are usually composed of RNA and DNA bases and the first two bases are always RNA. These primers are made by another enzyme called primase.
Every time a cell divides, DNA polymerase is required to help duplicate the cell’s DNA, so that a copy of the original DNA molecule can be passed to each of the daughter cells. In this way, genetic information is transmitted from generation to generation.
Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its tightly woven form. This opens up or “unzips” the double stranded DNA to give two single strands of DNA that can be used as templates for replication.
DNA polymerase adds new free nucleotides to the 3’ end of the newly-forming strand, elongating it in a 5’ to 3’ direction. However, DNA polymerase cannot begin the formation of this new chain on its own and can only add nucleotides to a pre-existing 3'-OH group. A primer is therefore needed, at which nucleotides can be added. Primers are usually composed of RNA and DNA bases and the first two bases are always RNA. These primers are made by another enzyme called primase.
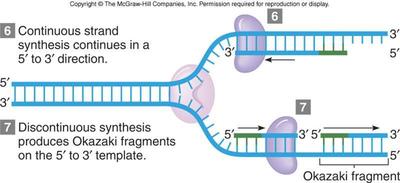
7.1 U 4 DNA replication is continuous on the leading strand and discontinuous on the lagging strand. (Oxford Biology Course Companion page 349).
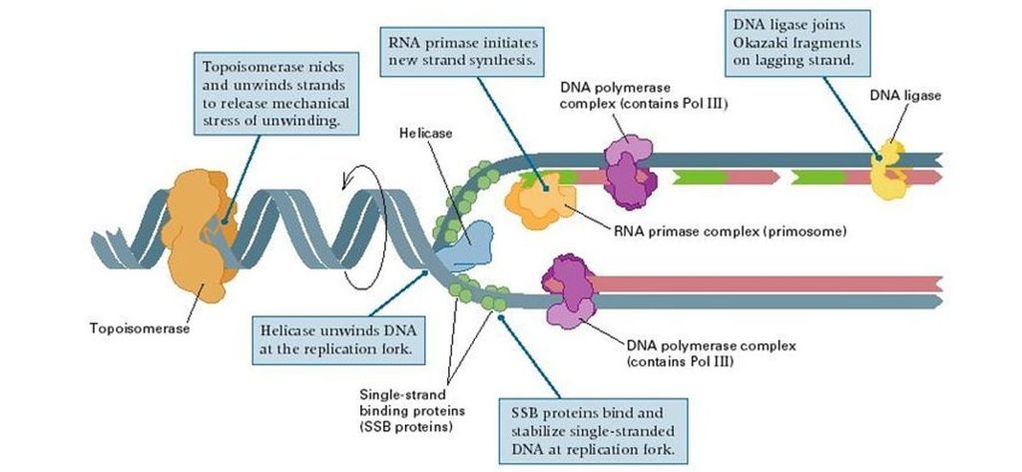
- Outline the role of the following proteins in DNA replications: helicase, topoisomerase (AKA gyrase), single stranded binding proteins, primase, DNA polymerase III, DNA polymerase I, and DNA ligase.
The DNA replication process is generally referred to as discontinuous, because the polymerizing enzyme can add nucleotides only in the 5' - 3' direction, synthesis is one strand (leading strand is continuous in the 5'-3' direction towards the fork. In the other strandt (lagging strand), as the forks opens, multiple sites of initiation are exposed. The synthesis, then proceed in short segments in the 5'-3' direction. That is , synthesis in the lagging strand is discontinuous. The short segments of DNA synthesized on the lagging strand are called Okazaki fragment. These fragments are about 1000 - 2000 nucleotides long
7.1 U 4 DNA replication is carried out by a complex system of enzymes. (Details of DNA replication differ between prokaryotes and eukaryotes. Only the prokaryotic system is expected. The proteins and enzymes involved in DNA replication should include helicase, DNA gyrase, single strand binding proteins, RNA primase and DNA polymerases I and III.
DNA replication creates two identical strands with each strand consisting of one new and one old strand (semi-conservative). DNA replication occurs at many different places on the DNA strand called the origins of replication (represented by bubbles along the strand).
- DNA gyrase:
- an enzyme that relieves strain while double-strand DNA is being unwound by helicase
- causes negative supercoiling of the DNA
- Helicase:
- controls unwinding of coiled DNA
- separates complementary strands of DNA, producing a replication fork
- single strand binding proteins:
- binds to single-stranded regions of DNA to prevent the two strands from rejoining by complementary base pairing
- allow other enzymes to function effectively on it
- RNA Primase:
- DNA polymerase III is only able to add DNA nucleotides to a free 3’ end on an existing DNA strand
- therefore, RNA primase uses the DNA template to synthesize a short 10 RNA nucleotide sequence known as an RNA primer
- DNA polymerase III:
- DNA polymerase III uses a single parent strand of DNA as a template
- adding free dexoyribonucleoside triphosphates from solution to the parent/template strand
- according to the complementary base pairing rules (A=T, G=C)
- DNA polymerase III can only add deoxyribonucleoside triphosphates to a free 3’ end of an existing nucleotide strand
- thus, on only one of the two strands of DNA can DNA polymerase III synthesize continuously in the direction toward the replication fork: this is known as the leading strand
- DNA polymerase I:
- DNA polymerase I is a proofreading enzyme
- removes the RNA nucleotides of the RNA primer
- replacing them with DNA nucleotides
- DNA ligase:
- forms covalent bonds linking together Okazaki fragments
- completing DNA synthesis along the lagging strand
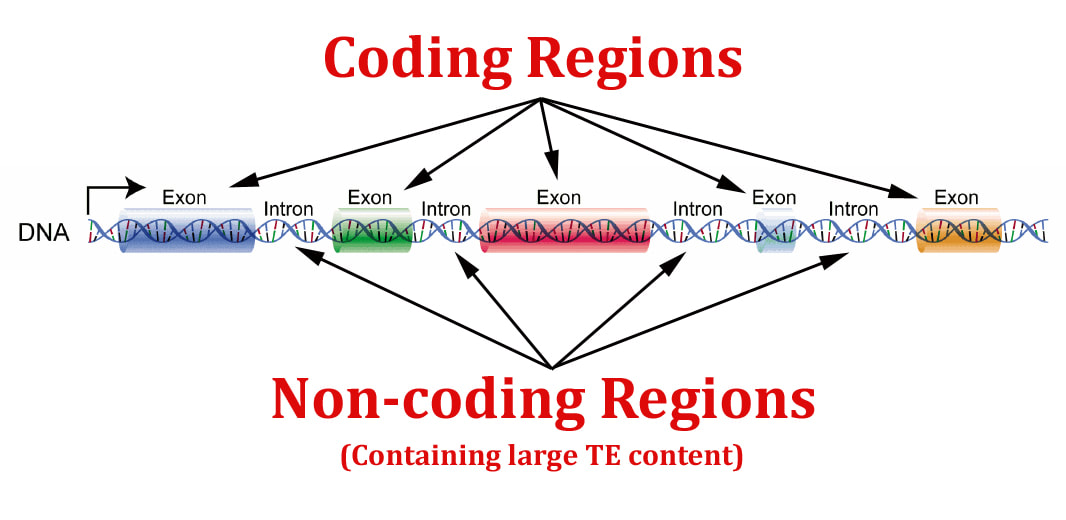
7.1 U 5 Some regions of DNA do not code for proteins but have other important functions. (Oxford Biology Course Companion page 350).
- Explain the need for RNA primers in DNA replication.
- Explain what is meant by DNA replication occurring in a 5' to 3' direction.
Genes contained within DNA called coding sequences, code for polypeptides created during transcription and translation. The majority of DNA are non-coding sequences that perform other functions such as regulators of gene expression, introns, telomeres and genes for tRNAs.
Unique or single-copy genes include exons and introns
Genes for other RNA types do not code for proteins
Some sections of DNA act as regulators of gene expression
Telomeres at the ends of chromosomes do not code for proteins
Highly repetitive sequences serve no known function
The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.
Unique or single-copy genes include exons and introns
- exons code for mature mRNA which codes for polypeptides
- introns are transcribed into RNA, but then removed by enzymes which splice together exons into mature mRNA, which codes for polypeptides
Genes for other RNA types do not code for proteins
- code for tRNA
- code for rRNA
Some sections of DNA act as regulators of gene expression
- regulators are involved in switching genes on or off
Telomeres at the ends of chromosomes do not code for proteins
Highly repetitive sequences serve no known function
- also known as satellite DNA, constitute 5-45% of the genome
- sequences are 5-300 base pairs per repeat, and my be repeated up to 10,000 times per genome
- the function of repetitive DNA is not known
- since repetitive sequences vary from person to person, they are useful in DNA profiling, which allows for DNA fingerprinting to identify samples from individuals
The regions of DNA that do not code for proteins should be limited to regulators of gene expression, introns, telomeres and genes for tRNAs.
- DNA sequencing of the human genome reveals that 98.5% does not code for proteins, rRNA or tRNA
- about a quarter of the human genome codes for introns and gene-related regulatory sequences
Applications
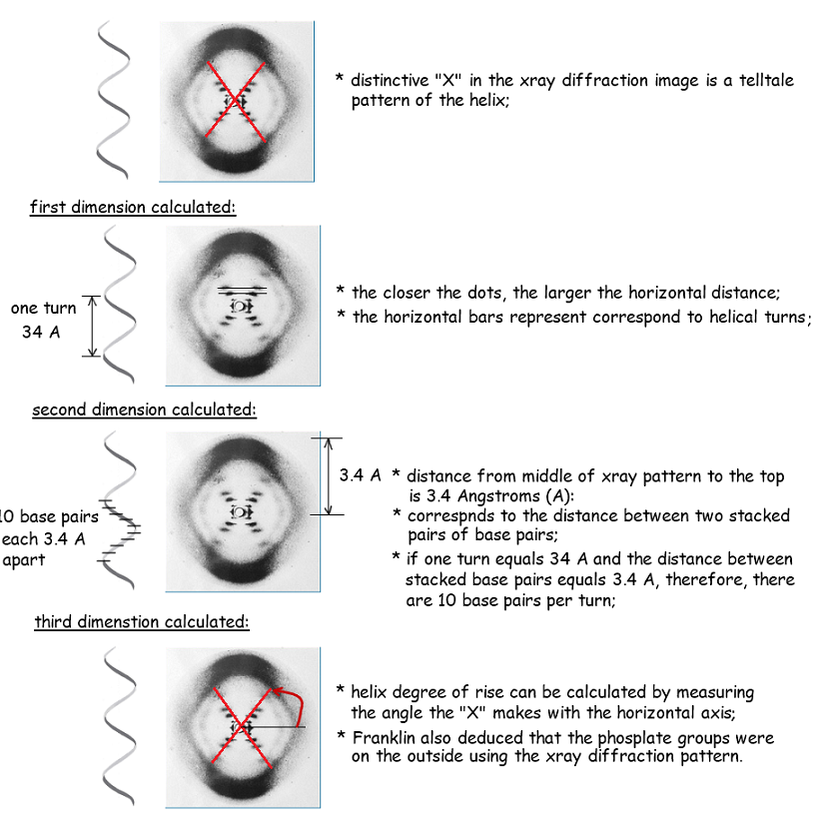
7.1 A 1 Rosalind Franklin’s and Maurice Wilkins’ investigation of DNA structure by X-ray diffraction. (Oxford Biology Course Companion page 346).
- Outline the process of X-ray diffraction.
- Outline the deductions about DNA structure made from the X-ray diffraction pattern.
Rosalind Franklin and Maurice Wilkins used a method of X-ray diffraction to investigate the structure of DNA. they were able to determine some helical dimensions using x-ray diffraction. X-ray diffraction patterns are regular, therefore, helix dimensions must be consistent.
Franklin’s data was shared by Wilkins with James Watson (without Franklin’s permission) who, with the help of Francise Crick, used the information to create a molecular model of the basic structure of DNA. In 1962, Watson, Crick and Wilkins (but not Franklin) were awarded the Nobel prize for their contributions to DNA structure identification
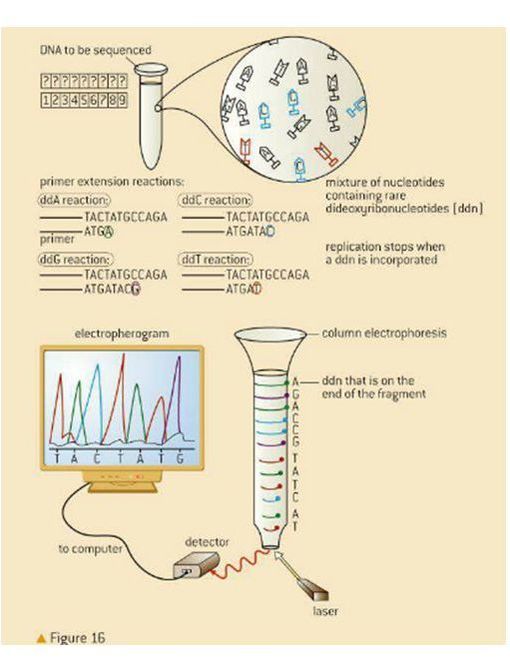
7.1 A 2 Use of nucleotides containing deoxyribonucleic acid to stop DNA replication in preparation of samples for base sequencing. (Oxford Biology Course Companion page 351).
- Define VNTR.
- Explain why VNTR are used in DNA profiling.
Dideoxyribonucleotides inhibit DNA polymerase during replication, thereby stopping replication from continuing. Dideoxyribonucleotides with fluorescent markers, are used and incorporated into sequences of DNA, to stop replication at the point at which they are added. This creates different sized fragments with fluorescent markers that can be separated by gel electrophoresis and analyzed by comparing the color of the fluorescence with the fragment length.
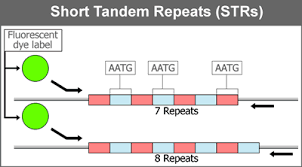
7.1 A 3 Tandem repeats are used in DNA profiling.
- Short tandem repeats (STRs), also known as variable tandem repeats (VNTRs) are regions of non-coding DNA that contain repeats of the same nucleotide sequence. These short repeats show variations between individuals in terms of the number of times the sequences is repeated.
- For example, CATACATACATACATACATACATA is a STR where the nucleotide sequence CATA is repeated six times for one individual. However, in another individual, this tandem repeat could occur only 4 times CATACATACATACATA. These variable tandem repeats are the basis for DNA profiling used in crime scene investigations and genealogical tests (paternity tests). The diagram below shows how the different number of these alleles for the VNTRs are used to create a DNA fingerprint of an individual.
Skill
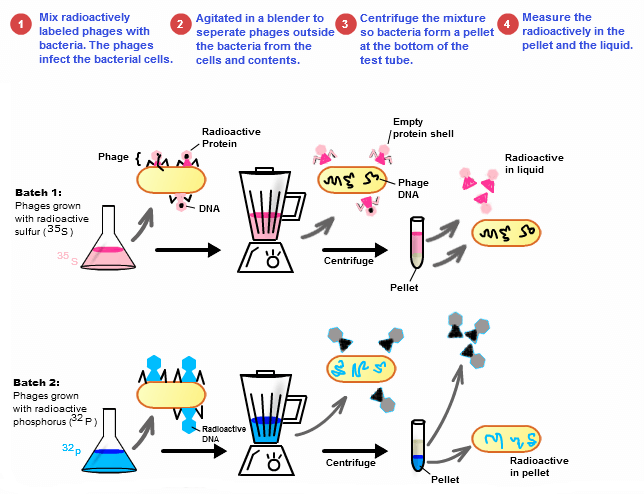
7.1 S 1 Analysis of results of the Hershey and Chase experiment providing evidence that DNA is the genetic material. (Oxford Biology Course Companion page 344).
- State the experimental question being tested in the Hershey and Chase experiment.
- Explain the procedure of the Hershey and Chase experiment.
- Explain how the results of the Hershey and Chase experiment supported the notion of nucleic acids as the genetic material.
In the mid-twentieth century, scientists were still unsure as to whether DNA or protein was the genetic material of the cell. It was known that some viruses consisted solely of DNA and a protein coat and could transfer their genetic material into hosts. In 1952, Alfred Hershey and Martha Chase conducted a series of experiments to prove that DNA was the genetic material
7.1 S 2 Utilization of molecular visualization software to analyse the association between protein and DNA within a nucleosome. (Oxford Biology Course Companion page 348)
- Identify nucleosome structures using molecular visualization software.
- Outline the mechanism of histone-DNA association.
COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT
Key Terms
|
H1 protein
Hershey-Chase leading strand DNA primase 3' DNA profile topoisomerase (AKA gyrase) Franklin-Wilkins single stranded binding proteins |
x-ray diffraction
lagging strand DNA polymerase I 5' short tandem supercoiling DNA ligase coding sequences |
nucleosomes
Okazaki fragments DNA polymerase III octamer core proteins hyper-variable repetitive sequences chain terminator nucleotides replication |
helicase
RNA primer parent DNA non-coding linker DNA primase telomere VNTR |
PowerPoint and Notes on Topic 7.1 by Chris Payne
Your browser does not support viewing this document. Click here to download the document.
Your browser does not support viewing this document. Click here to download the document.
Correct use of terminology is a key skill in Biology. It is essential to use key terms correctly when communicating your understanding, particularly in assessments. Use the quizlet flashcards or other tools such as learn, scatter, space race, speller and test to help you master the vocabulary.
,
Useful Links
Discovery of the Function of DNA Resulted from the Work of Multiple Scientists
DNA Is a Structure That Encodes Biological Information
In the News
Junk' DNA plays role in preventing breast cancer - ScienceDaily, Feb 2016
Researchers shed new light on regulation of repetitive DNA sequences - Phys.org, Jan 2016
Repetitive DNA provides hidden layer of functional information, researchers find - ScienceDaily, Dec 2015
Researchers Find Repetitive DNA Provides a Hidden Layer of Functional Information - NewWise, Dec 2015
Useful Links
Discovery of the Function of DNA Resulted from the Work of Multiple Scientists
DNA Is a Structure That Encodes Biological Information
In the News
Junk' DNA plays role in preventing breast cancer - ScienceDaily, Feb 2016
Researchers shed new light on regulation of repetitive DNA sequences - Phys.org, Jan 2016
Repetitive DNA provides hidden layer of functional information, researchers find - ScienceDaily, Dec 2015
Researchers Find Repetitive DNA Provides a Hidden Layer of Functional Information - NewWise, Dec 2015
TOK
- Highly repetitive sequences were once classified as “junk DNA” showing a degree of confidence that it had no role. To what extent do the labels and categories used in the pursuit of knowledge affect the knowledge we obtain?
Video Clips
Bill Nye talks about Greatest Discoveries
Nobel laureate James Watson opens TED2005 with the frank and funny story of how he and his research partner, Francis Crick, discovered the structure of DNA
Rosalind Franklin was a British scientist who helped discover the structure of DNA, but you most likely haven't heard of her. Hank will attempt to fix this gap in your knowledge on today's SciShow: Great Minds.
The discovery of the structure of the DNA double helix was one of the most important of the 20th century. In this educational video, explore Watson and Crick’s quest to understand DNA’s structure, and Rosalind Franklin’s key insights via x-ray crystallography.
Supercoiling of DNA
Short Tandem Repeats
Brief explanation of using STR for DNA Fingerprinting
How do we tell people apart by using their DNA? From murder investigations to paternity testing